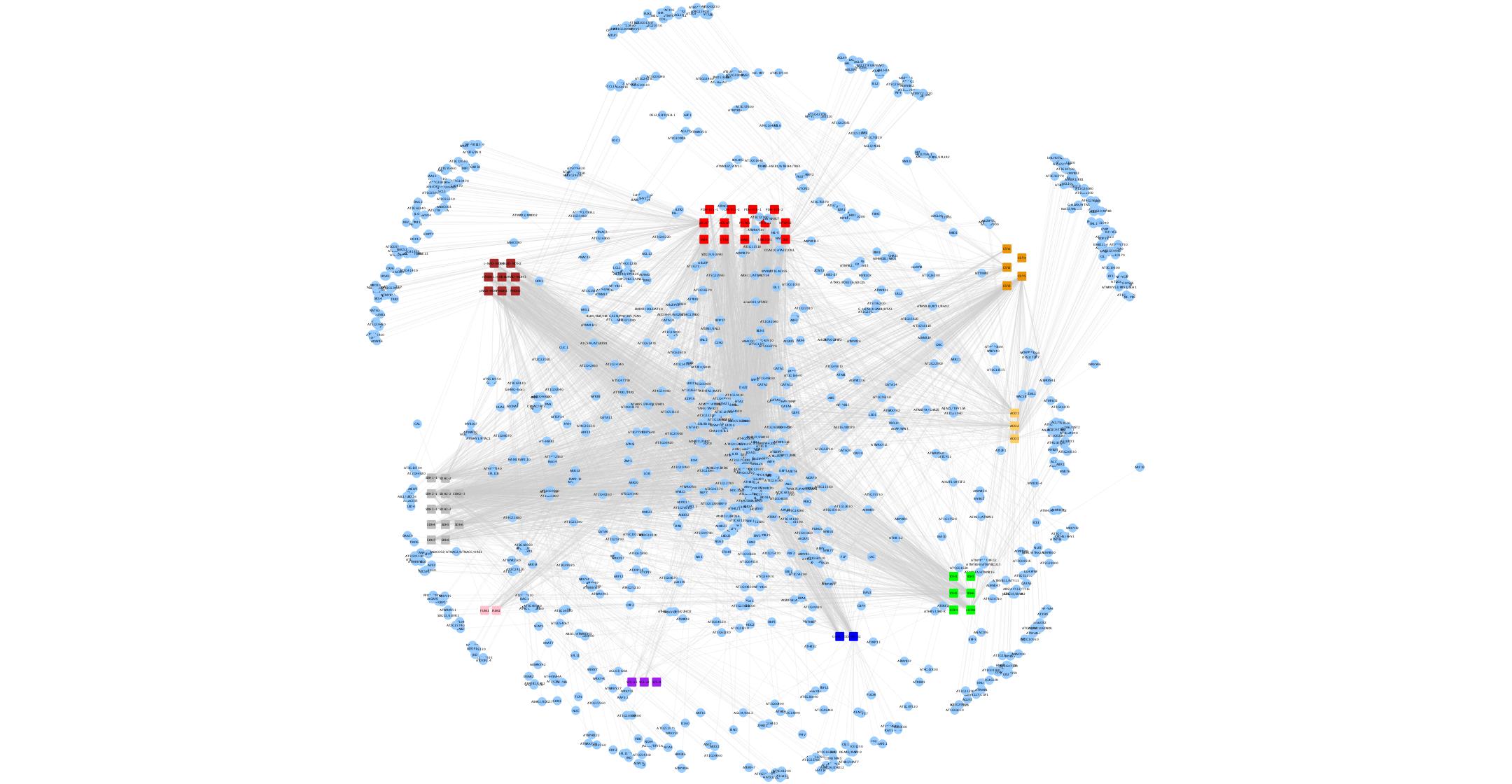
Primary metabolism functions to provide the energy and building blocks for life and reproduction. Even with this central role, very little is known about how primary metabolism is regulated, especially at the level of gene expression. This project will employ a systematic approach to develop models of how this regulation occurs. A series of experimental strategies is expected to identify a set of key regulatory modules that are predicted to be central for the control of primary metabolism. Mutants of select transcription factors will be used to assess the phenotypic consequences using a high-throughput platform. Physiological, metabolomic and transcriptional effects will be monitored to assess module function. The combined data generated will enable further experimental studies to test whether regulatory modules of primary metabolism are structured around specific biochemical networks, or whether they are structured around physiological outputs such as increased growth or yield. Intellectual Merit: Modern biology has a fundamental desire to understand how the genome of an organism programs its phenotype. However, this is greatly complicated by the need to move from the molecular to the macroscopic scale. It requires critical information of how transcriptional regulation and primary metabolism are interconnected and how regulatory modules connect at the molecular and physiological scales. Current biochemical and systems dogma says that gene regulation is structured around biosynthetic pathways; however this project will directly test an alternative hypothesis that modules have a more interconnected structure built around the physiological endpoints. Finally, this project will begin to develop a predictive model of how the genome governs the dynamics of primary metabolism.
Leadership:
PI: Daniel J. Kliebenstein UC Davis; co-PI Siobhan Brady (UC Davis)
Lab Memebers:
Michelle Tang
Funding:
NSF Molecular and Cellular Biology